A real-time qPCR identification and quantitation test
demonstrated the potential to be a valuable tool for measuring
severity of infection (and response to treatment).
David Farlow1,2, Lisa Simms1, Alex Pintara1, Graeme R. Nimmo3, Jacqueline Harper4, Karen Smith2, Matthew Hiskens2, Sarah Boxall2, Raffaella Giardino1, Anton Lord5, Flavia Huygens1
- Microbio Ltd., Brisbane, Australia
- Mackay Hospital and Health Service, Mackay, Australia
- School of Medicine, Griffith University, Gold Coast, Australia
- Pathology Queensland, Royal Brisbane and Women’s Hospital, Brisbane, Australia
- Specdata Consultants, Brisbane, Australia
Introduction
Sepsis is a common condition with high morbidity and mortality. Direct identification and quantitation of sepsis-associated pathogens has the potential to improve clinical decision making and targeted treatment of polymicrobial infections. Data from an Australian clinical observational study comparing traditional blood culture (BC) with InfectID-BSI, a direct from whole blood qPCR test, demonstrate the clinical value of rapid pathogen identification and quantitation in managing sepsis.
Methods
In total, 203 patients with suspected bloodstream infections from two Australian Emergency Departments were recruited into the study. Quantitation data and pathogen identification was available for 66 patients testing positive to a sepsis-associated pathogen using InfectID-BSI. Clinical concordance was based on three criteria: (i) severity of disease1; (ii) discharge diagnosis; (iii) identified pathogen, and classified as very likely, possible and unlikely. Less than 10 genome copies/0.35 mL blood (q<10) was considered a low bioburden2. Clinical metadata for 16 patients with q<10 were compared to 50 patients with q>10.
Results
1. Quantitation
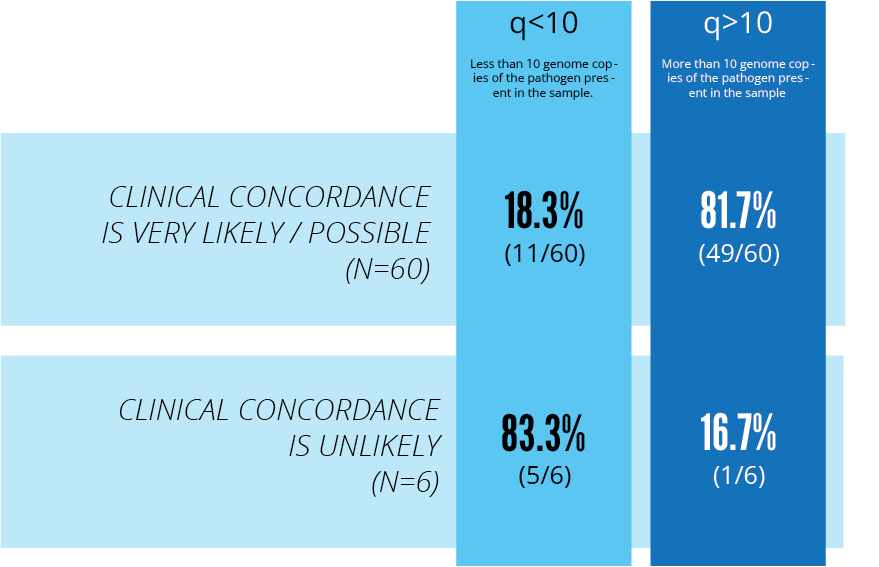
Quantitation data and pathogen identification was available for 66 patients. Clinical concordance (very likely/possible) was found for 60/66 (91%). Of the 60 patients, 49 patients had a q>10 (P value = 0.000397).
Of the 6/66 (9%) patients where clinical concordance with bloodstream infection/sepsis was unlikely, the majority had a q<10.
E. coli was the most prevalent pathogen identified and showed the highest bioburden where 17/66 (25%) of patients averaged q count of 3.228 x 103 with a 95% clinical concordance.
2. Time to positivity
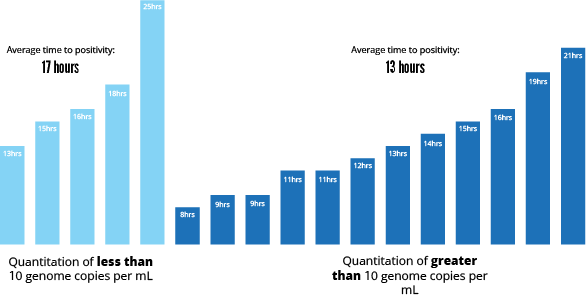
The average BC time to positivity (TTP) was 13.2 hours for patients with q>10 and 17.4 hours TTP with q<10.
3. Polymicrobial infections
For polymicrobial identifications, 0% of patients had q<10 but 32% (16/50) patients had q>10.
4. Malignancy comorbidities
For patients with malignancy comorbidities, 5/19 had q<10 and 14/19 had q>10.
Discussion
The InfectID-BSI real-time qPCR identification and quantitation assay has the potential to be a valuable tool for measuring severity of infection (and response to treatment). For patients with mixed infections, pathogen quantitation may assist clinical diagnosis and more precise patient treatment.
