Challenging Clinicians and Medical Microbiologists to reconsider sepsis management
Lisa Simms1, David Farlow1,3, Matthew I. Hiskens3, Tesfaye S. Mengistu3, Nicolas Sieben3, Brian McPherrin3, Janath De Silva3, Ron Nightingale3, Karen Smith3, Corey Davies1, Nadeesha Jayasundara1, Sumeet Sandhu1, Alex Pintara1, Raffaella Giardino1, Flavia Huygens*1
- Microbio Ltd., Brisbane, Australia
- Specdata Consultants, Brisbane, Australia
- Mackay Hospital and Health Service, Mackay, Australia
Introduction
Invasive pneumococcal disease and sepsis cause substantial morbidity and mortality worldwide resulting in long-term neurological sequelae in survivors, with case fatality rates of 11-30% in Europe and USA. Rapid molecular diagnostics provide clinicians with new sepsis management tools for patients with false negative blood culture results but with clinical signs and laboratory markers consistent with infection. Microbio’s novel sepsis diagnostic test, InfectID-BSI identifies and quantifies the top 26 sepsis causing bacteria and yeast in < 3 h directly from blood. In contrast, Blood Culture (BC) requires optimal conditions to grow pathogens. We report the incidence and quantitation of S. pneumoniae using InfectID-BSI.
Methods
A clinical observational study of 203 patients from two Australian Emergency Departments was undertaken. Patients were triaged using a sepsis risk assessment tool. Clinical and laboratory data fields were collected from electronic and paper-based medical records. Clinical concordance was examined within the context of commonly known body systems’ impact by individual pathogens, (eg pneumonia). Rare clinical impacts such as appendicitis, cellulitis and rhabdomyolysis relied on sparse literature review evidence of S. pneumoniae.
Results
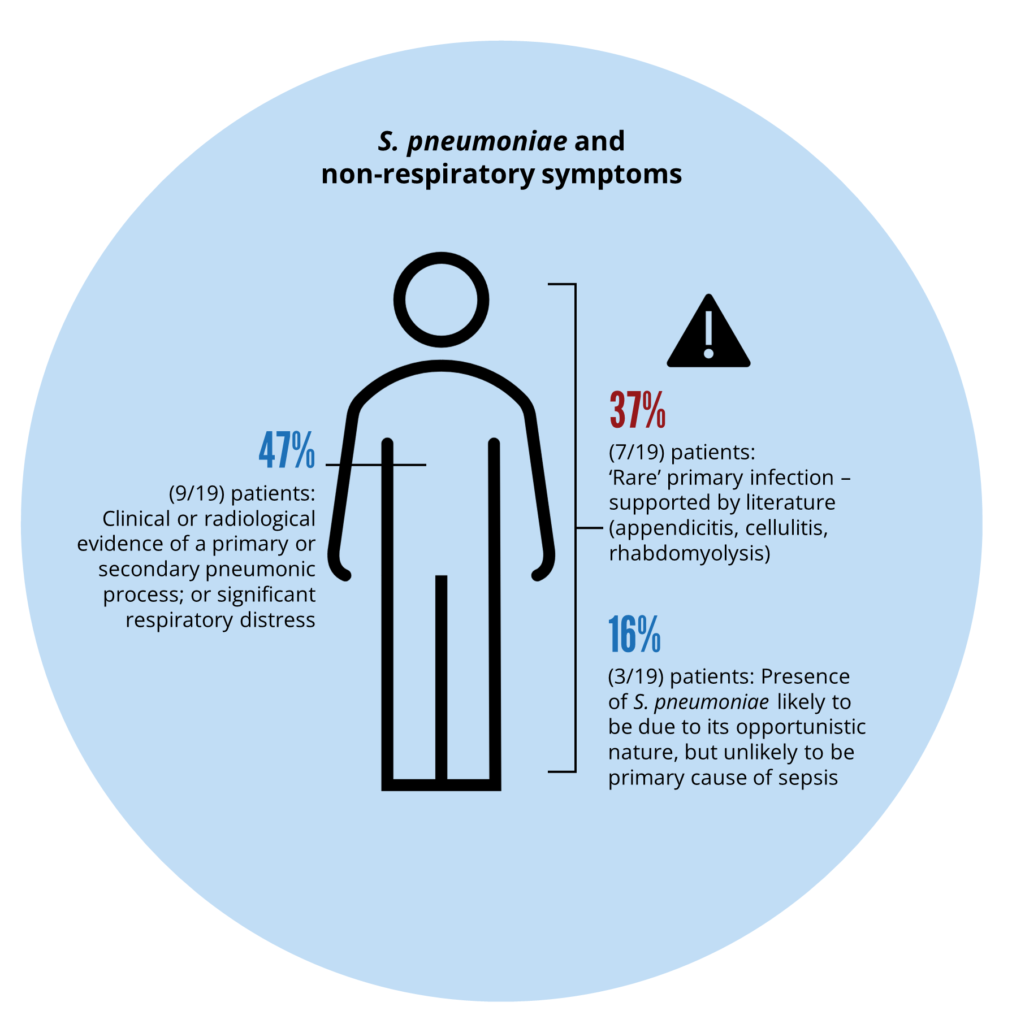
S. pneumoniae infection was detected in 19 patients using InfectID-BSI. Significantly, in each of these 19 patients, BC reported no growth. Nearly 50% of patients (9/19) with S. pneumoniae detections had clinical or radiological evidence of a pneumonic process or significant respiratory distress. Four of 19 (21.1%) patients had a diagnosis of urosepsis with Escherichia coli. Three of four patients had polymicrobial infection with low quantities of S. pneumoniae. Clinical concordance indicated that 80% of S. pneumoniae detections were likely the primary/secondary cause of sepsis. Rare clinical presentations (supported by the literature) included two patients with cellulitis, one with rhabdomyolysis, and one with appendicitis.
Table 1: An exemplar of polymicrobial infections with S. pneumoniae in four patients with a primary diagnosis of urosepsis, including Quantitation. S. pneumoniae infections were not detected by blood culture.
| Patient ID | Blood culture result | InfectID-BSI result [Quant: # genome copies / 0.35 mL blood] |
|---|---|---|
| MB5 | E. coli | E. coli [676*] S. pneumoniae [201] |
| MB13** | No growth | E. coli [676*] S. pneumoniae [201] |
| MB14 | No growth | E. coli [869] S. pneumoniae [95] |
| MB39** | No growth | E. coli [17,583*] S. pneumoniae [7] |
Discussion
InfectID-BSI assists clinicians in rapidly identifying S. pneumoniae, a fastidious pathogen. The quantitation tool assists in determining if the pathogen is the primary/secondary cause of sepsis in polymicrobial infections. Like all investigations (including blood culture), the results of InfectID-BSI must be used within the context of holistic clinical information.
