Simms, L.A., Davies, C., Jayasundara, N., Sandhu, S., Pintara, A., Pretorius, A., Nimmo, G.R., Harper, J., Hiskens, M., Smith, K., Boxall, S., Lord, A., Giardino, R., Farlow, D., Ward, D.M., Huygens, F., 2023. Performance evaluation of InfectID-BSI: A rapid quantitative PCR assay for detecting sepsis-associated organisms directly from whole blood. Journal of Microbiological Methods 211, 106783. Links:
https://doi.org/10.1016/j.mimet.2023.106783
Simms, L., Davies, C., Jayasundara, N., Sandhu, S., Pintara, A., Giardino, R., Lord, A., Huygens, F. (2022, April 23-26). A commercial innovative diagnostic assay for sepsis diagnosis directly from blood [Poster Presentation]. 32nd European Congress of Clinical Microbiology & Infectious Diseases 2022, Lisbon, Portugal
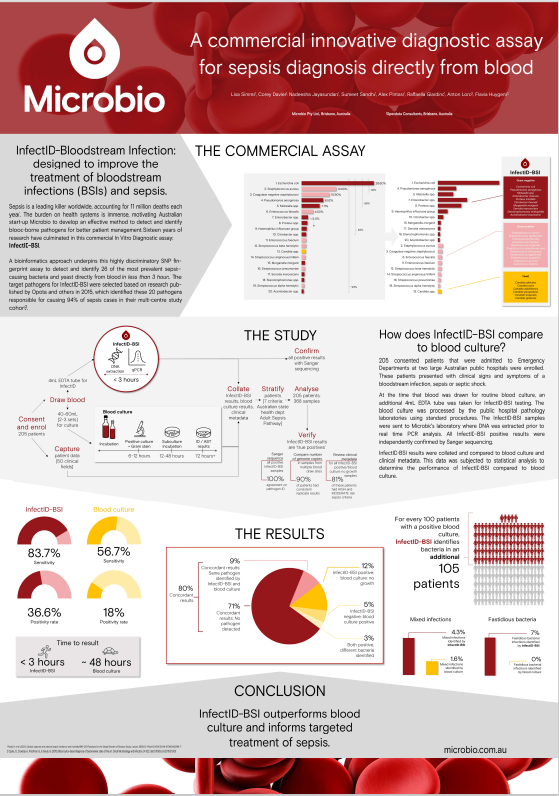
Farlow, D., Simms, L., Davies, C., Jayasundara, N., Sandhu, S., Pintara, A., Giardino, R., Lord, A., Huygens, F. (2022, April 23-26). A commercial assay versus traditional blood culture: correlation with clinical metadata and sepsis criteria [Poster Presentation]. 32nd European Congress of Clinical Microbiology & Infectious Diseases 2022, Lisbon, Portugal

Rathnayake, I., Hargreaves, M., & Huygens, F. (2011a). Genotyping of Enterococcus faecalis and Enterococcus faecium
isolates by use of a set of eight single nucleotide polymorphisms. Journal of Clinical Microbiology, 49, 367-72.Links:
https://jcm.asm.org/content/49/1/367
Rathnayake, I., Hargreaves, M., & Huygens, F. (2011b). SNP diversity of Enterococcus faecalis and Enterococcus faecium in
a South East Queensland waterway, Australia, and associated antibiotic resistance gene profiles. BMC Microbiology, 11,
201.Links:
https://bmcmicrobiol.biomedcentral.com/articles/10.1186/1471-2180-11-201
Sheludchenko, M., Huygens, F., & Hargreaves, M. (2011). Human-specific E. coli single nucleotide polymorphism (SNP)
genotypes detected in a South East Queensland waterway, Australia. Environmental Science and Technology, 45,
10331-6.Links:
https://pubs.acs.org/doi/10.1021/es201599u10.1021/es201599u
Sheludchenko, M., Huygens, F., & Hargreaves, M. (2010). Highly discriminatory single-nucleotide polymorphism
interrogation of Escherichia coli by use of allele-specific real-time PCR and eBURST analysis. Applied and Environmental
Microbiology, 76, 4337-45.Links:
https://aem.asm.org/content/76/13/433710.1128/AEM.00128-10
Honsa, E., Fricke, R., Stephens, A., Ko, D., Kong, F., Gilbert, G., Huygens, F., Giffard, P. (2008). Assignment of Streptococcus
agalactiae isolates to aclonal complexes using a small set of single nucleotide polymorphisms. BMC Microbiology, 19(8),
140.Links:
https://bmcmicrobiol.biomedcentral.com/articles/10.1186/1471-2180-8-14010.1186/1471-2180-8-140
Huygens, F., Inman-Bamber, J. N., Munckhof, W., Schooneveldt, J., Harrison, B., McMahon, J., & Giffard, P. (2006).
Staphylococcus aureus genotyping using real-time PCR formats. Journal of Clinical Microbiology, 44, 3712-9.Links:
https://jcm.asm.org/content/44/10/371210.1128/JCM.00843-06
Price, E., Thiruvenkataswamy, V., Mickan, L., Unicomb, L., Rios, R., Huygens, F., & Giffard, P. (2006). Genotyping of
Campylobacter jejuni using seven single-nucleotide polymorphisms in combination with flaA short variable region
sequencing. Journal of Medical Microbiology, 55, 1061-70.Links:
https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.46460-010.1099/jmm.0.46460-0
Stephens, A., Huygens, F., Inman-Bamber, J., Price, E. N., Schooneveldt, J., Munckhof, W., & Giffard, P. (2006). Methicillin-
resistant Staphylococcus aureus genotyping using a small set of polymorphisms. Journal of Medical Microbiology, 55,
43-51.Links:
https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.46157-0
Robertson, G., Thiruvenkataswamy, V., Shilling, H., Price, E., Huygens, F. H., & Giffard, P. (2004). Identification and
interrogation of highly informative single nucleotide polymorphism sets defined by bacterial multilocus sequence
typing databases. Journal of Medical Microbiology, 53, 35-45.Links:
https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.05365-0
